Novel microfluidic method for bioseparation
 Researchers at the Brown University have devised a simple new technique that can separate tiny amounts of the target molecules from biological samples by single motion of a magnet under a microchannel. The novel microfluidic method is important and broadly applicable, especially in biological diagnostic platforms that require binding and separation of known target biomolecules, such as RNA, single stranded DNA (ssDNA) or proteins.
Researchers at the Brown University have devised a simple new technique that can separate tiny amounts of the target molecules from biological samples by single motion of a magnet under a microchannel. The novel microfluidic method is important and broadly applicable, especially in biological diagnostic platforms that require binding and separation of known target biomolecules, such as RNA, single stranded DNA (ssDNA) or proteins.
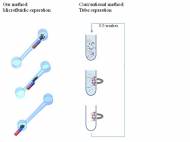
The separation of target molecules from biological samples is a crucial part of today’s diagnostics and detection strategies. The target and probe molecules are mixed and then separated in a batch process that requires multiple pipetting, tube washing and extraction steps that can decrease accuracy. The novel technique may replace pipettes and test tubes in some diagnostic applications and increase the accuracy and sensitivity of disease detection.
The microfluidic platform, developed by Anubhav Tripathi and his team at Brown, utilizes a mobile magnetic field in static microfluidic channels, where ssDNA molecules are isolated via nucleic acid hybridization. This platform is static and it doesn’t rely on external pumps to mix samples or flow target molecules. Research team modified bead-like magnetic particles by attaching short pieces of DNA to them in order to capture target DNA molecules with specific sequences matching. DNA molecules are then separated by pulling the magnetic particles along the channel.
The process is simple, fast and highly specific. It is applicable for point-of-care platforms that are used to detect bacterial, viral infections and prion diseases by DNA, RNA or protein identification. According to the researchers, specific disease applications include testing for HIV and influenza. Also, the technique can be used to estimate the expression of certain protein markers, such as troponin (an indicator of damage to the heart muscle) or any detection that requires binding and separation of known target biomolecules.
The biggest challenge for the Tripathi’s team was optimizing the system and characterizing the chip for biological assay because it required that both engineering as well as biological factors need to be considered. However, assays that use this platform are already developed.
The research team has developed a new microchip based Simple Method of Amplifying RNA Targets (SMART) assay that detects influenza from patient samples. SMART assay matches the results of Polymerase Chain Reaction (PCR), which is the most used method for influenza diagnosis. The team’s next challenge is developing essays using the microfluidic platform to detect wild type and drug-resistant HIV in areas with limited resources such as Kenya and South Africa.
For more information, read the paper published in the journal Biomicrofluidics: “Microfluidic Platform for Isolating Nucleic Acid Targets Using Sequence Specific Hybridization“.









Leave your response!